Learning Objectives
By the end of this section, you will be able to:
- Describe horizontal gene transfer
- Illustrate how prokaryotes and eukaryotes transfer genes horizontally
- Identify the web and ring models of phylogenetic relationships and describe how they differ from the original phylogenetic tree concept
The concepts of phylogenetic modeling are constantly changing. It is one of the most dynamic fields of study in all of biology. Over the last several decades, new research has challenged scientists’ ideas about how organisms are related. New models of these relationships have been proposed for consideration by the scientific community.
Many phylogenetic trees have been shown as models of the evolutionary relationship among species. Phylogenetic trees originated with Charles Darwin, who sketched the first phylogenetic tree in 1837 ([Figure 1]a), which served as a pattern for subsequent studies for more than a century. The concept of a phylogenetic tree with a single trunk representing a common ancestor, with the branches representing the divergence of species from this ancestor, fits well with the structure of many common trees, such as the oak ([Figure 1]b). However, evidence from modern DNA sequence analysis and newly developed computer algorithms has caused skepticism about the validity of the standard tree model in the scientific community.
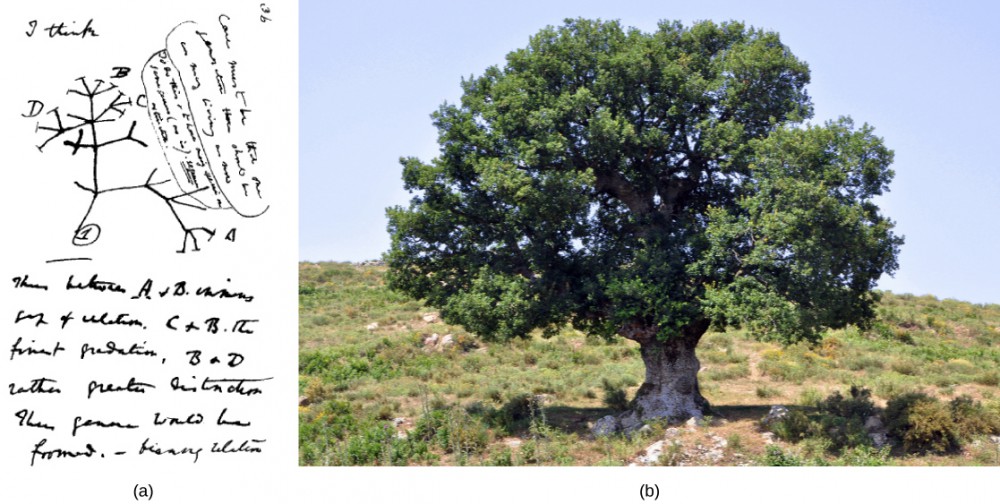
Figure 1: The (a) concept of the “tree of life” goes back to an 1837 sketch by Charles Darwin. Like an (b) oak tree, the “tree of life” has a single trunk and many branches. (credit b: modification of work by “Amada44″/Wikimedia Commons)
Limitations to the Classic Model
Classical thinking about prokaryotic evolution, included in the classic tree model, is that species evolve clonally. That is, they produce offspring themselves with only random mutations causing the descent into the variety of modern-day and extinct species known to science. This view is somewhat complicated in eukaryotes that reproduce sexually, but the laws of Mendelian genetics explain the variation in offspring, again, to be a result of a mutation within the species. The concept of genes being transferred between unrelated species was not considered as a possibility until relatively recently. Horizontal gene transfer (HGT), also known as lateral gene transfer, is the transfer of genes between unrelated species. HGT has been shown to be an ever-present phenomenon, with many evolutionists postulating a major role for this process in evolution, thus complicating the simple tree model. Genes have been shown to be passed between species which are only distantly related using standard phylogeny, thus adding a layer of complexity to the understanding of phylogenetic relationships.
The various ways that HGT occurs in prokaryotes is important to understanding phylogenies. Although at present HGT is not viewed as important to eukaryotic evolution, HGT does occur in this domain as well. Finally, as an example of the ultimate gene transfer, theories of genome fusion between symbiotic or endosymbiotic organisms have been proposed to explain an event of great importance—the evolution of the first eukaryotic cell, without which humans could not have come into existence.
Horizontal Gene Transfer
Horizontal gene transfer (HGT) is the introduction of genetic material from one species to another species by mechanisms other than the vertical transmission from parent(s) to offspring. These transfers allow even distantly related species to share genes, influencing their phenotypes. It is thought that HGT is more prevalent in prokaryotes, but that only about 2% of the prokaryotic genome may be transferred by this process. Some researchers believe such estimates are premature: the actual importance of HGT to evolutionary processes must be viewed as a work in progress. As the phenomenon is investigated more thoroughly, it may be revealed to be more common. Many scientists believe that HGT and mutation appear to be (especially in prokaryotes) a significant source of genetic variation, which is the raw material for the process of natural selection. These transfers may occur between any two species that share an intimate relationship ([Figure 1]).
| Summary of Mechanisms of Prokaryotic and Eukaryotic HGT | |||
|---|---|---|---|
| Mechanism | Mode of Transmission | Example | |
| Prokaryotes | transformation | DNA uptake | many prokaryotes |
| transduction | bacteriophage (virus) | bacteria | |
| conjugation | pilus | many prokaryotes | |
| gene transfer agents | phage-like particles | purple non-sulfur bacteria | |
| Eukaryotes | from food organisms | unknown | aphid |
| jumping genes | transposons | rice and millet plants | |
| epiphytes/parasites | unknown | yew tree fungi | |
| from viral infections | |||
HGT in Prokaryotes
The mechanism of HGT has been shown to be quite common in the prokaryotic domains of Bacteria and Archaea, significantly changing the way their evolution is viewed. The majority of evolutionary models, such as in the Endosymbiont Theory, propose that eukaryotes descended from multiple prokaryotes, which makes HGT all the more important to understanding the phylogenetic relationships of all extant and extinct species.
The fact that genes are transferred among common bacteria is well known to microbiology students. These gene transfers between species are the major mechanism whereby bacteria acquire resistance to antibiotics. Classically, this type of transfer has been thought to occur by three different mechanisms:
- Transformation: naked DNA is taken up by a bacteria
- Transduction: genes are transferred using a virus
- Conjugation: the use a hollow tube called a pilus to transfer genes between organisms
More recently, a fourth mechanism of gene transfer between prokaryotes has been discovered. Small, virus-like particles called gene transfer agents (GTAs) transfer random genomic segments from one species of prokaryote to another. GTAs have been shown to be responsible for genetic changes, sometimes at a very high frequency compared to other evolutionary processes. The first GTA was characterized in 1974 using purple, non-sulfur bacteria. These GTAs, which are thought to be bacteriophages that lost the ability to reproduce on their own, carry random pieces of DNA from one organism to another. The ability of GTAs to act with high frequency has been demonstrated in controlled studies using marine bacteria. Gene transfer events in marine prokaryotes, either by GTAs or by viruses, have been estimated to be as high as 1013 per year in the Mediterranean Sea alone. GTAs and viruses are thought to be efficient HGT vehicles with a major impact on prokaryotic evolution.
As a consequence of this modern DNA analysis, the idea that eukaryotes evolved directly from Archaea has fallen out of favor. While eukaryotes share many features that are absent in bacteria, such as the TATA box (found in the promoter region of many genes), the discovery that some eukaryotic genes were more homologous with bacterial DNA than Archaea DNA made this idea less tenable. Furthermore, the fusion of genomes from Archaea and Bacteria by endosymbiosis has been proposed as the ultimate event in eukaryotic evolution.
HGT in Eukaryotes
Although it is easy to see how prokaryotes exchange genetic material by HGT, it was initially thought that this process was absent in eukaryotes. After all, prokaryotes are but single cells exposed directly to their environment, whereas the sex cells of multicellular organisms are usually sequestered in protected parts of the body. It follows from this idea that the gene transfers between multicellular eukaryotes should be more difficult. Indeed, it is thought that this process is rarer in eukaryotes and has a much smaller evolutionary impact than in prokaryotes. In spite of this fact, HGT between distantly related organisms has been demonstrated in several eukaryotic species, and it is possible that more examples will be discovered in the future.
In plants, gene transfer has been observed in species that cannot cross-pollinate by normal means. Transposons or “jumping genes” have been shown to transfer between rice and millet plant species. Furthermore, fungal species feeding on yew trees, from which the anti-cancer drug TAXOL® is derived from the bark, have acquired the ability to make taxol themselves, a clear example of gene transfer.
In animals, a particularly interesting example of HGT occurs within the aphid species ([Figure 2]). Aphids are insects that vary in color based on carotenoid content. Carotenoids are pigments made by a variety of plants, fungi, and microbes, and they serve a variety of functions in animals, who obtain these chemicals from their food. Humans require carotenoids to synthesize vitamin A, and we obtain them by eating orange fruits and vegetables: carrots, apricots, mangoes, and sweet potatoes. On the other hand, aphids have acquired the ability to make the carotenoids on their own. According to DNA analysis, this ability is due to the transfer of fungal genes into the insect by HGT, presumably as the insect consumed fungi for food. A carotenoid enzyme called a desaturase is responsible for the red coloration seen in certain aphids, and it has been further shown that when this gene is inactivated by mutation, the aphids revert back to their more common green color ([Figure 2]).

Figure 2: (a) Red aphids get their color from red carotenoid pigment. Genes necessary to make this pigment are present in certain fungi, and scientists speculate that aphids acquired these genes through HGT after consuming fungi for food. If genes for making carotenoids are inactivated by mutation, the aphids revert back to (b) their green color. Red coloration makes the aphids a lot more conspicuous to predators, but evidence suggests that red aphids are more resistant to insecticides than green ones. Thus, red aphids may be more fit to survive in some environments than green ones. (credit a: modification of work by Benny Mazur; credit b: modification of work by Mick Talbot)
Genome Fusion and the Evolution of Eukaryotes
Scientists believe the ultimate in HGT occurs through genome fusion between different species of prokaryotes when two symbiotic organisms become endosymbiotic. This occurs when one species is taken inside the cytoplasm of another species, which ultimately results in a genome consisting of genes from both the endosymbiont and the host. This mechanism is an aspect of the Endosymbiont Theory, which is accepted by a majority of biologists as the mechanism whereby eukaryotic cells obtained their mitochondria and chloroplasts. However, the role of endosymbiosis in the development of the nucleus is more controversial. Nuclear and mitochondrial DNA are thought to be of different (separate) evolutionary origin, with the mitochondrial DNA being derived from the circular genomes of bacteria that were engulfed by ancient prokaryotic cells. Mitochondrial DNA can be regarded as the smallest chromosome. Interestingly enough, mitochondrial DNA is inherited only from the mother. The mitochondrial DNA degrades in sperm when the sperm degrades in the fertilized egg or in other instances when the mitochondria located in the flagellum of the sperm fails to enter the egg.
Within the past decade, the process of genome fusion by endosymbiosis has been proposed by James Lake of the UCLA/NASA Astrobiology Institute to be responsible for the evolution of the first eukaryotic cells ([Figure 3]a). Using DNA analysis and a new mathematical algorithm called conditioned reconstruction (CR), his laboratory proposed that eukaryotic cells developed from an endosymbiotic gene fusion between two species, one an Archaea and the other a Bacteria. As mentioned, some eukaryotic genes resemble those of Archaea, whereas others resemble those from Bacteria. An endosymbiotic fusion event, such as Lake has proposed, would clearly explain this observation. On the other hand, this work is new and the CR algorithm is relatively unsubstantiated, which causes many scientists to resist this hypothesis.
More recent work by Lake ([Figure 3]b) proposes that gram-negative bacteria, which are unique within their domain in that they contain two lipid bilayer membranes, indeed resulted from an endosymbiotic fusion of archaeal and bacterial species. The double membrane would be a direct result of the endosymbiosis, with the endosymbiont picking up the second membrane from the host as it was internalized. This mechanism has also been used to explain the double membranes found in mitochondria and chloroplasts. Lake’s work is not without skepticism, and the ideas are still debated within the biological science community. In addition to Lake’s hypothesis, there are several other competing theories as to the origin of eukaryotes. How did the eukaryotic nucleus evolve? One theory is that the prokaryotic cells produced an additional membrane that surrounded the bacterial chromosome. Some bacteria have the DNA enclosed by two membranes; however, there is no evidence of a nucleolus or nuclear pores. Other proteobacteria also have membrane-bound chromosomes. If the eukaryotic nucleus evolved this way, we would expect one of the two types of prokaryotes to be more closely related to eukaryotes.
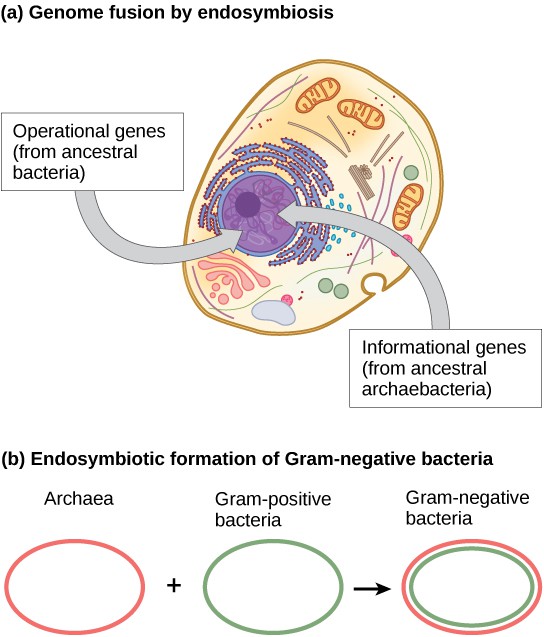
Figure 3: The theory that mitochondria and chloroplasts are endosymbiotic in origin is now widely accepted. More controversial is the proposal that (a) the eukaryotic nucleus resulted from the fusion of archaeal and bacterial genomes, and that (b) Gram-negative bacteria, which have two membranes, resulted from the fusion of Archaea and Gram-positive bacteria, each of which has a single membrane.
The nucleus-first hypothesis proposes that the nucleus evolved in prokaryotes first ([Figure 4]a), followed by a later fusion of the new eukaryote with bacteria that became mitochondria. The mitochondria-first hypothesis proposes that mitochondria were first established in a prokaryotic host ([Figure 4]b), which subsequently acquired a nucleus, by fusion or other mechanisms, to become the first eukaryotic cell. Most interestingly, the eukaryote-first hypothesis proposes that prokaryotes actually evolved from eukaryotes by losing genes and complexity ([Figure 4]c). All of these hypotheses are testable. Only time and more experimentation will determine which hypothesis is best supported by data.
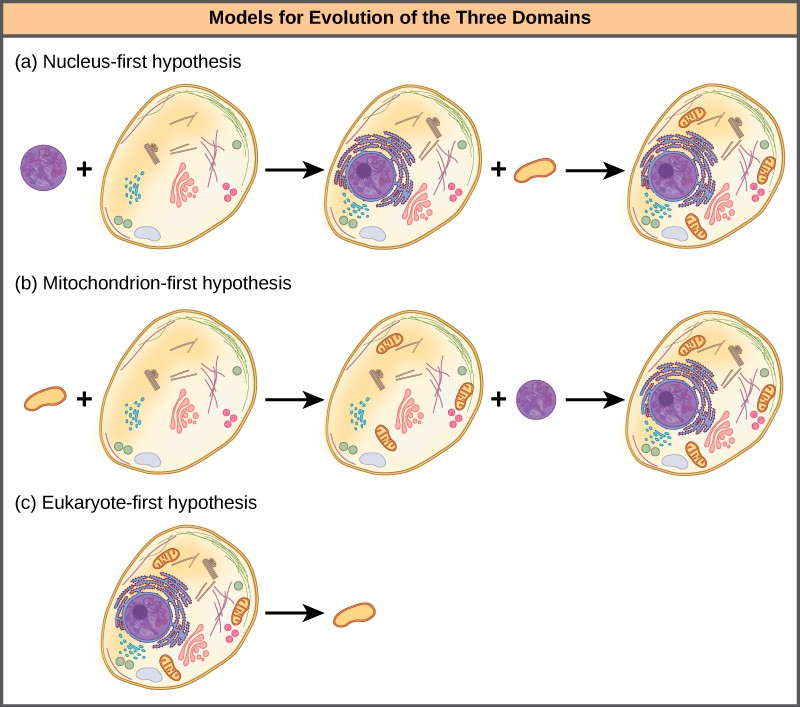
Figure 4: Three alternate hypotheses of eukaryotic and prokaryotic evolution are (a) the nucleus-first hypothesis, (b) the mitochondrion-first hypothesis, and (c) the eukaryote-first hypothesis.
Web and Network Models
The recognition of the importance of HGT, especially in the evolution of prokaryotes, has caused some to propose abandoning the classic “tree of life” model. In 1999, W. Ford Doolittle proposed a phylogenetic model that resembles a web or a network more than a tree. The hypothesis is that eukaryotes evolved not from a single prokaryotic ancestor, but from a pool of many species that were sharing genes by HGT mechanisms. As shown in [Figure 5]a, some individual prokaryotes were responsible for transferring the bacteria that caused mitochondrial development to the new eukaryotes, whereas other species transferred the bacteria that gave rise to chloroplasts. This model is often called the “web of life.” In an effort to save the tree analogy, some have proposed using the Ficus tree ([Figure 5]b) with its multiple trunks as a phylogenetic to represent a diminished evolutionary role for HGT.
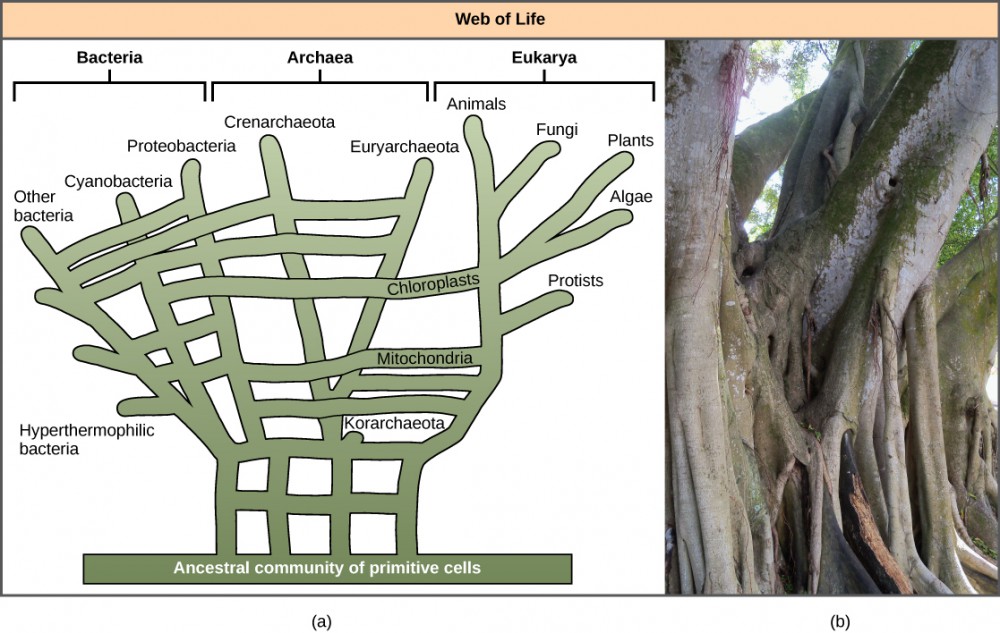
Figure 5: In the (a) phylogenetic model proposed by W. Ford Doolittle, the “tree of life” arose from a community of ancestral cells, has multiple trunks, and has connections between branches where horizontal gene transfer has occurred. Visually, this concept is better represented by (b) the multi-trunked Ficus than by the single trunk of the oak similar to the tree drawn by Darwin [Figure 1]. (credit b: modification of work by “psyberartist”/Flickr)
Ring of Life Models
Others have proposed abandoning any tree-like model of phylogeny in favor of a ring structure, the so-called “ring of life” ([Figure 6]); a phylogenetic model where all three domains of life evolved from a pool of primitive prokaryotes. Lake, again using the conditioned reconstruction algorithm, proposes a ring-like model in which species of all three domains—Archaea, Bacteria, and Eukarya—evolved from a single pool of gene-swapping prokaryotes. His laboratory proposes that this structure is the best fit for data from extensive DNA analyses performed in his laboratory, and that the ring model is the only one that adequately takes HGT and genomic fusion into account. However, other phylogeneticists remain highly skeptical of this model.
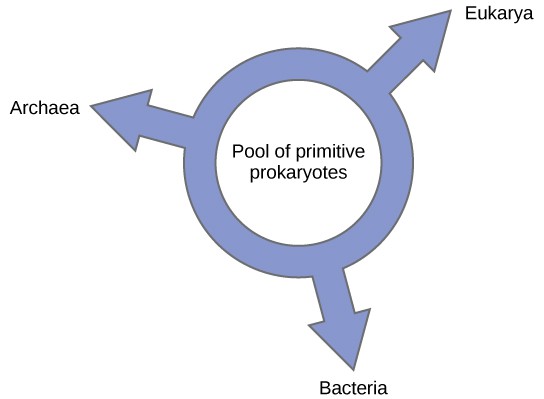
Figure 6: According to the “ring of life” phylogenetic model, the three domains of life evolved from a pool of primitive prokaryotes.
In summary, the “tree of life” model proposed by Darwin must be modified to include HGT. Does this mean abandoning the tree model completely? Even Lake argues that all attempts should be made to discover some modification of the tree model to allow it to accurately fit his data, and only the inability to do so will sway people toward his ring proposal.
This doesn’t mean a tree, web, or a ring will correlate completely to an accurate description of phylogenetic relationships of life. A consequence of the new thinking about phylogenetic models is the idea that Darwin’s original conception of the phylogenetic tree is too simple, but made sense based on what was known at the time. However, the search for a more useful model moves on: each model serving as hypotheses to be tested with the possibility of developing new models. This is how science advances. These models are used as visualizations to help construct hypothetical evolutionary relationships and understand the massive amount of data being analyzed.
Section Summary
The phylogenetic tree, first used by Darwin, is the classic “tree of life” model describing phylogenetic relationships among species, and the most common model used today. New ideas about HGT and genome fusion have caused some to suggest revising the model to resemble webs or rings.
Review Questions
The transfer of genes by a mechanism not involving asexual reproduction is called:
- meiosis
- web of life
- horizontal gene transfer
- gene fusion
Particles that transfer genetic material from one species to another, especially in marine prokaryotes:
- horizontal gene transfer
- lateral gene transfer
- genome fusion device
- gene transfer agents
What does the trunk of the classic phylogenetic tree represent?
- single common ancestor
- pool of ancestral organisms
- new species
- old species
Which phylogenetic model proposes that all three domains of life evolved from a pool of primitive prokaryotes?
- tree of life
- web of life
- ring of life
- network model
Free Response
Compare three different ways that eukaryotic cells may have evolved.
Describe how aphids acquired the ability to change color.
Glossary
- eukaryote-first hypothesis
- proposal that prokaryotes evolved from eukaryotes
- gene transfer agent (GTA)
- bacteriophage-like particle that transfers random genomic segments from one species of prokaryote to another
- genome fusion
- fusion of two prokaryotic genomes, presumably by endosymbiosis
- horizontal gene transfer (HGT)
- (also, lateral gene transfer) transfer of genes between unrelated species
- mitochondria-first hypothesis
- proposal that prokaryotes acquired a mitochondrion first, followed by nuclear development
- nucleus-first hypothesis
- proposal that prokaryotes acquired a nucleus first, and then the mitochondrion
- ring of life
- phylogenetic model where all three domains of life evolved from a pool of primitive prokaryotes
- web of life
- phylogenetic model that attempts to incorporate the effects of horizontal gene transfer on evolution
Candela Citations
- Biology. Provided by: OpenStax CNX. Located at: http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8. License: CC BY: Attribution. License Terms: Download for free at http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8
- Introduction to Ecology & Evolutionary Biology, Derived from Biology by OpenStax. Authored by: Adrienne Correa. Located at: http://cnx.org/contents/25adc78c-f54a-4ef4-a081-ebe711c480ed@3.19. License: CC BY: Attribution. License Terms: Download for free at http://cnx.org/contents/25adc78c-f54a-4ef4-a081-ebe711c480ed@3.19
