Project Goals and Objectives
The Human Skin Microbiome Project is an application of the principles and practices of classic microbiological investigation. During this project, which will take several weeks to complete, you will survey the microbial inhabitants of the human skin (yours). Then, you will apply an expanding array of microbiology laboratory skills to grow and investigate the colonial, cellular, and metabolic properties of one of the bacterial species from your skin culture.
More specifically, you will:
- prepare a primary culture from your skin, and observe the colonial and cellular properties of the bacteria that grow on it;
- identify one skin isolate that you would like to investigate further, and maintain it in a pure culture for an extended period of time;
- use microbiological methods to investigate the cellular and metabolic properties of your skin isolate;
- understand basic principles of taxonomy and how to apply the information in Bergey’s Manual of Determinative Bacteriology to presumptively identify your skin isolate.
Biosafety Considerations
While it may seem somewhat ironic that bacteria you have been carrying around on your skin forever are now going to be classified as a potential biohazard and subjected to risk assessment and laboratory containment practices, it is nonetheless an important consideration.
Of the various types of bacteria that might be encountered during the primary culture stage of this project, most are BSL-1, and some BSL-2. To minimize the risks of working with bacteria of unknown identity, this project will be limited to only Gram positive bacteria, and BSL-2 practices will be employed while working with cultures of your isolate.
Bergey’s Manual
The definitive reference book for bacterial taxonomy and identification is Bergey’s Manual. There are two versions: the manual of Systematic Bacteriology, which is concerned with issues of bacterial taxonomy and systematics (arranging bacteria into taxa according to similarities/differences in DNA), and the manual of Determinative Bacteriology, which deals specifically with the identification aspects of bacterial taxonomy. The latter book will be our primary resource for this project. For an overview of the manual and how to use it, read Chapters I, II, III, IV, and V.
The Human Skin Microbiome
The bacteria and other microbes that live on human skin are those that are best adapted to survive the prevailing conditions. Regions of the human body can be thought of as different ecosystems. Exposed, dry areas of the skin, such as the forearm, are akin to an arid desert environment, which is a preferred environment for many Gram-positive bacteria. Skin sites that are generally dark, warm, and moist, such as the underarm or perineum, are similar to temperate or tropical ecosystems; they tend to harbor more microbes in general and are more likely to have a larger percentage of Gram-negative bacteria.
The quantitative differences found at these sites may relate to the amount of moisture, body temperature, and varying concentrations of skin surface lipids. Most microorganisms live in the superficial layers of the skin (the stratum corneum) and in the upper parts of the hair follicles. Some bacteria, however, reside in the deeper areas of the hair follicles, where they may be beyond the reach of ordinary disinfection procedures (like washing your face with soap/water or an antibacterial product). These out-of-reach bacteria serve as a reservoir for recolonization of the skin environment after the surface bacteria are removed.
Figure 1 illustrates the types of bacteria that are commonly found on various regions of the human skin. Not all of these bacteria are culturable, because the growth conditions necessary for their survival are difficult to replicate in an artificial environment. Using the culture conditions established for this laboratory, the bacteria grown in your primary culture will most likely be Actinobacteria (Micrococcus, Corynebacterium, Mycobacterium), Firmicutes (Staphylococcus or other Gram-positive bacteria), or Proteobacteria (Gram negative bacteria).

Figure 1. Types of bacteria found on human skin
The skin microbiome may include fungi such as yeasts and molds as well as bacteria. While interesting, these eukaryotic microorganisms are outside of the scope of this project. Molds form very distinctive colonies that will be easy to identify as fungal in origin, and thus, easy to avoid. Yeasts, however, produce colonies that resemble those of bacteria, although typically smaller and different in color. When you select the colony and make a pure culture, avoid colonies that have the appearance of either a mold (furry, fuzzy, or powdery) or a yeast (very small, very slow growing—only appearing after a week or more of incubation, and brightly colored—red, orange, pink, or even bright white colonies).
Over the course of several weeks, you will maintain your bacterial strain in pure culture while performing tests to determine its colonial, cellular, and metabolic properties, and ultimately its “presumptive” identity. The term “presumptive” is used because phenotypic methods are less exact than those that rely on a direct analysis of DNA.
Record all observations, test outcomes, and interpretations on the Human Skin Microbiome Project Worksheets, according to the instructions provided by your instructor.
Identification of an Unknown Bacterium
Bergey’s Manual contains an enormous amount of information about the characteristics of all known bacterial species, mostly presented in table form. You will use the information in these tables to determine the identity of your skin isolate. To aid in this process, and as a way to demonstrate how you ruled out all other possible bacterial species, a taxonomic tool called a dichotomous key (also called a diagnostic key or sequential key) will be used to narrow down the possibilities.
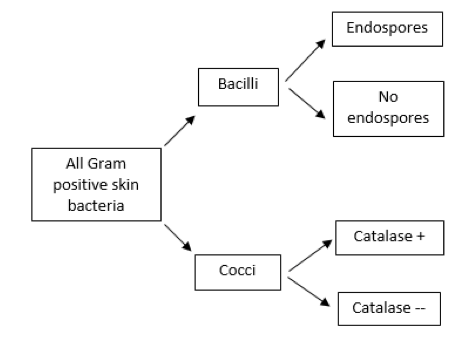
Figure 2. Example of a dichotomous key
A dichotomous key is a sequence of questions with two possible mutually exclusive answers (a “couplet,” such as yes or no, positive or negative, cocci or bacilli). Starting with a large group of bacteria (in this case, all of the possible Gram-positive bacteria that can be found on the human skin), the first question should relate to an observable characteristic for which there are two choices, followed by additional questions until the possibilities are narrowed to a single choice. A brief example is shown in Figure 2.
Bergey’s Manual of Determinative Bacteriology is designed to facilitate identification, and not classification, of bacteria based primarily on phenotypic observations. In Bergey’s, the broadest grouping is represented by the four Major Categories. The primary criterion to establish the Major Category for your skin microbe will be the nature of its cell wall. Remember that you are being limited to choosing a bacterium that is Gram positive; therefore, the starting point for this project will be bacteria that are in Major Category II: Gram-positive eubacteria that have cell walls. Because of the culture methods and conditions we will employ in this project, it is highly unlikely that your isolate would be from Categories III (eubacteria lacking cell walls) or IV (archaeobacteria); thus, those are excluded.
At this point, you have narrowed your options down from all bacteria to only those classified as Category II. The next step is to identify the Group, within the Major Category, to which your isolate belongs. Chapter V in Bergey’s provides a list and brief summary of the bacteria in the Groups within each Category; Table V.2 refers specifically to Category II bacteria. Cellular morphology (for example, cell shape and presence of endospores) and physiology (particularly with regard to your isolate’s physiological oxygen requirements and relative results) will help you decide on a Group. Once you made that decision, you can follow the directions given in Table V.2 to find the first page of the identification tables in Bergey’s.
Continue to narrow your choices down to a single Genus within the Group, and once you’ve decided on a genus, locate and refer to the identification table that specifies the different Species within that genus. The tables also list the laboratory tests routinely performed to differentiate among the species, and the expected outcomes of those tests for each bacterial species in the genus.
You may not be able to conduct all of the tests listed in Bergey’s. Different tests may be available for you to perform, depending on the laboratory in which you work. For the purpose of identifying your skin microbe, below is a list of the tests that are available for you to use in the ID process.
- Gram stain and endospore stain
- 7.5% salt tolerance (MSA)
- Mannitol fermentation (MSA for salt-tolerant bacteria)
- Bile tolerance and esculin hydrolysis (BE)
- Hemolysis on blood agar
- Glucose fermentation (MR; VP; TSI)
- Lactose/sucrose fermentation (TSI)
- Fermentation of other carbohydrates (see instructor)
- Susceptibility or resistance to antibiotics
- Acetoin production (VP)
- Catalase
- Oxidase
- Production of H2S (TSI; SIM)
- Indole (SIM)
- Motility (SIM)
- Citrate utilization
- Nitrate reduction
- Coagulase (for staphylococci ONLY)
- Urease
Other tests may be available, as indicated by your instructor.
Observations, Outcomes, and Next Steps
The following tasks will be performed over the course of several weeks:
Project Step 1: Make a primary culture from your skin on a TSA plate and incubate it for up to a week at room temperature.
Project Step 2: After incubation, the TSA plate that contains the primary culture from your skin will very likely hold many, hopefully well isolated, colonies.
From among these, choose 3 colonies for subculture and further examination. Remember to avoid colonies that appear to be a mold (typically large, green or brown, and fuzzy) or yeast (very small and vibrantly white or a shade of red). With a sterile inoculation loop, subculture each colony to a section of a TSA plate, to create a pure culture, as you did previously and illustrated in Figure 3. Incubate the plate at RT until bacterial growth is abundant.

Figure 3. Subcultures on different sections of a TSA plate
Project Step 3: Gram stain each of the three pure cultures. Choose one that is Gram-positive (bacilli or cocci) as your project bacterium (your skin microbe, or HSM). Subculture your HSM to a TSA slant and incubate it. Once there is abundant growth on the surface, store the TSA slant culture in the refrigerator. Make sure the slant is clearly labelled with your name.
Complete the Colonial and Cellular Morphology (CCM) Worksheet.
Project Step 4: From the observations you’ve made and the results of lab tests,use Bergey’s Manual as a reference to determine the Major Category, Group and then Genus of your HSM.
Complete the Genus Identification (GID) Worksheet.
Project Step 5: Once you’ve narrowed your options down to a single genus, locate the specific table in Bergey’s Manual for identification of individual species. Based on the information in the table, compile a list of appropriate tests that will facilitate species identification. Select those that are available for you to perform, as specified by your lab instructor. Perform the appropriate tests and record the results.
Project Step 6: Compare your results with the expected test outcomes from Bergey’s Manual to determine the Species of your human skin microbe.
Complete the Species Identification (SID) Worksheet.
HSMP Worksheet 1: Colonial and Cellular Morphology
Name __________________________________________ Date due _________________
Record the observations/results obtained so far below. NOTE: your instructor may make this worksheet available to you electronically through a course management system, and may request that you type your answers into the worksheet before printing and handing it in.
1. Name the region of your body from which you obtained the specimen for the primary culture.
2. Based on colony appearance, approximately how many different types of bacteria from your skin are represented on the TSA streak plate of your primary culture?
3. Based on the appearance of an isolated colony and using appropriate microbiology terminology, describe the colonial morphology of each of the three bacterial subcultures.
| Colony Type 1 | ||
| Colony size | ||
| Texture | ||
| Transparency | ||
| Pigmentation | ||
| Form (shape, margin, elevation) | ||
| Colony Type 2 | ||
| Colony size | ||
| Texture | ||
| Transparency | ||
| Pigmentation | ||
| Form (shape, margin, elevation) | ||
| Colony Type 3 | ||
| Colony size | ||
| Texture | ||
| Transparency | ||
| Pigmentation | ||
| Form (shape, margin, elevation) | ||
4. For each of the three bacterial pure cultures, describe the outcome of the Gram stain using appropriate microbiology terminology:
| Gram Stain Outcome | Cell shape | Arrangement | |
| Colony 1 | |||
| Colony 2 | |||
| Colony 3 |
5. Of the three bacteria you investigated, choose one that is Gram-positive as your project bacterium. Below, indicate which of the three you chose and restate the Gram staining result.
Colony # and Gram stain result (including cell shape and arrangement):
_____________________________________________________________________
6. Compare and contrast the chemical composition and structure of the cell wall of a Gram-positive bacterium such as your isolate, with the cell wall of a Gram negative bacterium.
7. Briefly discuss why Gram-positive cells appear purple, and Gram-negative cells appear pink, after the Gram stain process is applied.
8. Briefly discuss how bacterial cells produce “arrangements” that we can observe with a microscope.
9. State whether it will be necessary for you to perform an endospore stain on your isolate, and give a specific reason to explain why you should, or should not, use this staining method to identify your isolate.
10. Briefly explain why it is necessary to include a mixture of iodine and potassium iodide (Gram’s Iodine) in the overall Gram stain procedure.
In addition to this worksheet, you may also be asked to prepare and provide to your instructor a Gram stained slide of a smear prepared from your environmental isolate to evaluate your technique. If evaluated, the following criteria will be used: single layer of cells is achieved, cell morphology and arrangement are easily determined, all cells appear the same color, shape, and arrangement, and there are no visible contaminants.
Instructor Evaluation of Gram Stained Slide:
Gram stain result as observed by instructor: _______________________________________
Evaluation of technique: _______________________________________________________
Criteria: Single layer of cells is achieved; cell morphology and arrangement are easily determined; all cells appear the same color, shape, and arrangement; and there are no visible contaminants. Degree of concurrence with instructor’s description of cellular morphology will also be noted.
HSMP Worksheet 2: Genus Identification
Name __________________________________________ Date due _________________
1. From your CCM worksheet, restate the Gram stain results (reaction, morphology, and arrangement) for your skin isolate:
2. Taxonomic Classification: State the Domain and Phylum for your isolate, based on the evidence you have accumulated so far.
3. Report the results of the following physiological tests performed on your environmental isolate:
| Test | Describe in detail the outcomes of the following tests (meaning, what you directly observe: such as bubbles after H2O2 was added; red color on the slant and yellow on the butt with cracks; etc.) | Interpretation of observed outcome (for example; pos or neg; K/A, gas, etc) |
| Catalase | ||
| Oxidase | ||
| TSI agar | ||
| Nitrate Reduction |
4. From your observations of bacterial growth characteristics and physiological tests up to this point, state ALL energy metabolism pathway(s) used by your skin bacterium to make ATP. Then provide convincing evidence from among your observations and test results to support your determination.
| Energy Metabolism pathway | Do your observations and/or results of the tests above indicate that your EI uses this pathway? (YES or NO) | STATE one observation and/or test result that provides scientific evidence for that pathway, and explain why/how the result indicates that your EI bacterium uses this pathway. |
| Aerobic respiration | ||
| Anaerobic respiration | ||
| Fermentation |
5. Growth Category for Oxygen
Based on the observed growth patterns and test results above, state the physiological oxygen requirement (strict aerobe, microaerophile, strict anaerobe, facultative anaerobe, or aerotolerant anaerobe) for your EI bacterium.
6. Bergey’s Group
Table V in Bergey’s is divided into four sections, one for each Major Category of bacteria. Remember that you were directed to select a Gram-positive bacterium as your EI, so look at Table V.2 to determine to which Group, within Major Category II, your EI should be assigned. Based on the characteristics of your EI bacterium that you have observed up to this point:
(a) State the Group (group number and name) for your EI according to the Bergey’s Manual identification system.
(b)State TWO observations that provide scientific evidence to support your choice of Group designation from (a) above.
7. Tests to assign Genus
If necessary (and it may not be necessary at this point), perform additional tests to determine the genus of your environmental isolate. Describe those tests and their outcomes in the table below. If you can determine the genus without additional tests, don’t put anything in this table.
| Test/Observation | Describe in detail the outcomes of the following tests (meaning, what you directly observe): such as bubbles after H2O2 was added; red color on the slant and yellow on the butt with cracks; etc.) | Interpretation of observed outcome (for example; pos or neg; K/A, gas, etc) |
8. Genus ID Flowchart (dichotomous key)
A brief example of how to construct a dichotomous key was provided previously in this lab. Note that the key you develop will be used by your instructor to review the process and logic of your choice of genus.
Some advice on how to proceed: Remember that your goal is to RULE OUT genera that are not consistent with the characteristics you’ve observed for your EI bacterium. Start by listing ALL of the possible genera in the Bergey’s Group. Then look at the characteristics that distinguish the various genera from one another (such as cell shape, endospore production, growth on human skin, etc.). As your first couplet, choose a feature that your isolate exhibits and the majority of other genera lack. Then, for the genera that were not ruled out, choose as the next couplet a feature that again rules out as many genera as possible. Continue until there is only a single genus left that is consistent with all of your observations made up to this point.
State the presumptive genus of your EI bacterium: ____________________________________
HSMP Worksheet 3: Species Identification
Name ____________________________________________Date Due _______________
1. Review of observations/characteristics of your EI determined so far:
| Pigmentation of colonies (color ONLY) | |
| Gram stain reaction | |
| Cellular morphology | |
| Cellular arrangement | |
| Endospores observed (yes or no) | |
| Result of catalase test (+ or -) | |
| Result of the oxidase test (+ or -) | |
| Result of TSI | |
| Result of nitrate reduction test (+ or -) | |
| List ALL energy pathways indicated | |
| Growth category for oxygen | |
| Bergey’s Group (# and name) | |
| Name of Genus |
2. Tests for Species ID
Locate the specific Table in Bergey’s Manual that shows the species within the genus and the tests needed for the differentiation and identification of your isolate. Below, list the tests you will need to perform (in addition to those already done) to presumptively identify your isolate. Cross reference the list of tests available in your laboratory (provided previously by your instructor).
3. Complete the table below with the tests/outcomes you observed for your EI bacterium. The number of rows in the table is arbitrary – only do as many tests as needed to ID your isolate. Add additional rows, if necessary).
| Name of test | Direct observation of the outcome of test(s) (how did the media, slide, tube, etc. APPEAR when you looked at it?) | Outcome/Result (pos or neg, K/A, etc.) |
4. As you did previously, construct a dichotomous key to show the process and logic used to presumptively identify your environmental isolate. Begin by listing ALL species within the genus, below. Note that subspecies (if there are any) should be listed individually.
5. Complete EITHER A or B below:
A. If you were able to identify a single species after completing all possible tests: Write the full binomial name (using scientific nomenclature) for your environmental isolate below:
B. If you were NOT able to discriminate a single species after completing all your tests: Write the full binomial name of ALL remaining species and provide the reason why you were unable to assign a single species to your EI.
6. Growth Characteristics
Use biology/microbiology terminology to state the specific category related to the following growth characteristics for your EI. Provide your reasoning for the choice of each category, including specific examples of growth patterns observed for your isolate from among your observations and tests.
| Physiological Category | Supporting evidence from among your observations/test results | |
| Nutritional | ||
| Temperature | ||
| Osmotic (salt) tolerance |
7. Fully classify your isolate by providing the following information, using appropriate terminology and scientific nomenclature:
| Taxon | Classification for your skin isolate |
| Domain | |
| Phylum | |
| Class | |
| Order | |
| Family | |
| Genus | |
| Species |
