Learning Outcomes
- Understand the difference between pre-RNA and pre-mRNA
After transcription, eukaryotic pre-mRNAs must undergo several processing steps before they can be translated. Eukaryotic (and prokaryotic) tRNAs and rRNAs also undergo processing before they can function as components in the protein synthesis machinery.
mRNA Processing
The eukaryotic pre-mRNA undergoes extensive processing before it is ready to be translated. The additional steps involved in eukaryotic mRNA maturation create a molecule with a much longer half-life than a prokaryotic mRNA. Eukaryotic mRNAs last for several hours, whereas the typical E. coli mRNA lasts no more than five seconds.
The three most important steps of pre-mRNA processing are the addition of stabilizing and signaling factors at the 5′ and 3′ ends of the molecule, and the removal of intervening sequences that do not specify the appropriate amino acids.
5′ Capping
A cap is added to the 5′ end of the growing transcript by a phosphate linkage. This addition protects the mRNA from degradation. In addition, factors involved in protein synthesis recognize the cap to help initiate translation by ribosomes.
3′ Poly-A Tail
Once elongation is complete, an enzyme called poly-A polymerase adds a string of approximately 200 A residues, called the poly-A tail to the pre-mRNA. This modification further protects the pre-mRNA from degradation and signals the export of the cellular factors that the transcript needs to the cytoplasm.
Pre-mRNA Splicing
Eukaryotic genes are composed of exons, which correspond to protein-coding sequences (ex-on signifies that they are expressed), and intervening sequences called introns (intron denotes their intervening role), which are removed from the pre-mRNA during processing. Intron sequences in mRNA do not encode functional proteins.
All of a pre-mRNA’s introns must be completely and precisely removed before protein synthesis. If the process errs by even a single nucleotide, the reading frame of the rejoined exons would shift, and the resulting protein would be dysfunctional. The process of removing introns and reconnecting exons is called splicing (Figure 1).
Practice Question
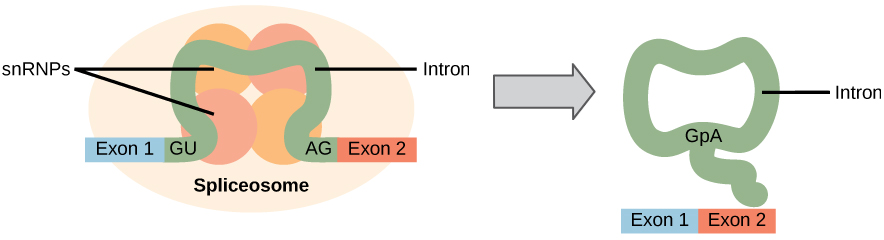
Figure 1. Pre-mRNA splicing involves the precise removal of introns from the primary RNA transcript. The splicing process is catalyzed by protein complexes called spliceosomes that are composed of proteins and RNA molecules called snRNAs. Spliceosomes recognize sequences at the 5′ and 3′ end of the intron.
Errors in splicing are implicated in cancers and other human diseases. What kinds of mutations might lead to splicing errors?
Try It
Candela Citations
- Biology. Provided by: OpenStax CNX. Located at: http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8. License: CC BY: Attribution. License Terms: Download for free at http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8
