Learning Outcomes
- Understand RNA splicing and explain its role in regulating gene expression
- Describe the importance of RNA stability in gene regulation
RNA is transcribed, but must be processed into a mature form before translation can begin. This processing after an RNA molecule has been transcribed, but before it is translated into a protein, is called post-transcriptional modification. As with the epigenetic and transcriptional stages of processing, this post-transcriptional step can also be regulated to control gene expression in the cell. If the RNA is not processed, shuttled, or translated, then no protein will be synthesized.
RNA splicing, the first stage of post-transcriptional control
In eukaryotic cells, the RNA transcript often contains regions, called introns, that are removed prior to translation. The regions of RNA that code for protein are called exons (Figure 1). After an RNA molecule has been transcribed, but prior to its departure from the nucleus to be translated, the RNA is processed and the introns are removed by splicing.
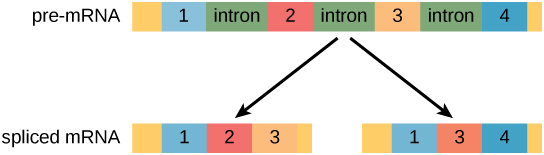
Figure 1. Pre-mRNA can be alternatively spliced to create different proteins.
Alternative RNA Splicing
Alternative RNA splicing is a mechanism that allows different protein products to be produced from one gene when different combinations of introns, and sometimes exons, are removed from the transcript (Figure 2).
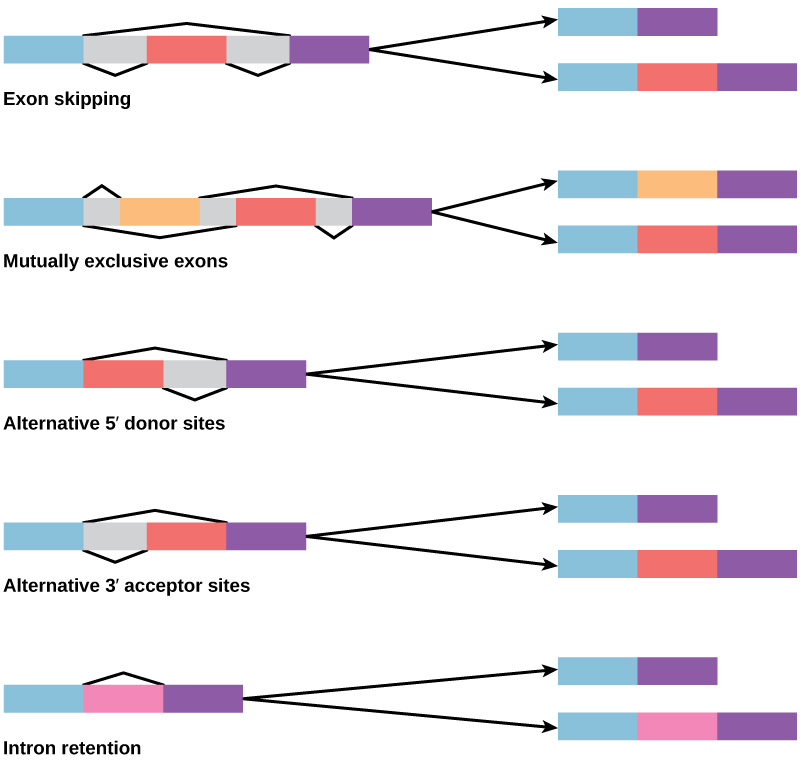
Figure 2. There are five basic modes of alternative splicing.
This alternative splicing can be haphazard, but more often it is controlled and acts as a mechanism of gene regulation.
Visualize how mRNA splicing happens by watching the process in action in this video:
Control of RNA Stability
Before the mRNA leaves the nucleus, it is given two protective “caps” that prevent the end of the strand from degrading during its journey. The 5′ cap, which is placed on the 5′ end of the mRNA and poly-A tail, which is attached to the 3′ end. Once the RNA is transported to the cytoplasm, the length of time that the RNA resides there can be controlled. Each RNA molecule has a defined lifespan and decays at a specific rate. This rate of decay is referred to as the RNA stability. If the RNA is stable, it will be detected for longer periods of time in the cytoplasm.
RNA Stability and microRNAs
The microRNAs, or miRNAs, are short single-stranded RNA molecules that are only 21–24 nucleotides in length. Like transcription factors and RBPs, mature miRNAs recognize a specific sequence and bind to the RNA to degrade the target mRNA. They rapidly destroy the RNA molecule.
In Summary: Post-Transcriptional Control of Gene Expression
Post-transcriptional regulation can occur at any stage after transcription, including RNA splicing, nuclear shuttling, and RNA stability. Once RNA is transcribed, it must be processed to create a mature RNA that is ready to be translated. This involves the removal of introns that do not code for protein. Spliceosomes bind to the signals that mark the exon/intron border to remove the introns and ligate the exons together. Once this occurs, the RNA is mature and can be translated. RNA is created and spliced in the nucleus, but needs to be transported to the cytoplasm to be translated. RNA is transported to the cytoplasm through the nuclear pore complex. Once the RNA is in the cytoplasm, the length of time it resides there before being degraded, called RNA stability, can also be altered to control the overall amount of protein that is synthesized. RNA stability is controlled by microRNAs (miRNAs). These miRNAs bind to the 5′ CAP or the 3′ Tail of the RNA to decrease RNA stability and promote decay.
Practice Questions
Which of the following are involved in post-transcriptional control?
- control of RNA splicing
- control of RNA shuttling
- control of RNA stability
- all of the above
Binding of a miRNAs will ________ the stability of the RNA molecule.
- increase
- decrease
- neither increase nor decrease
- either increase or decrease
Try It
Candela Citations
- Biology. Provided by: OpenStax CNX. Located at: http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8. License: CC BY: Attribution. License Terms: Download for free at http://cnx.org/contents/185cbf87-c72e-48f5-b51e-f14f21b5eabd@10.8
